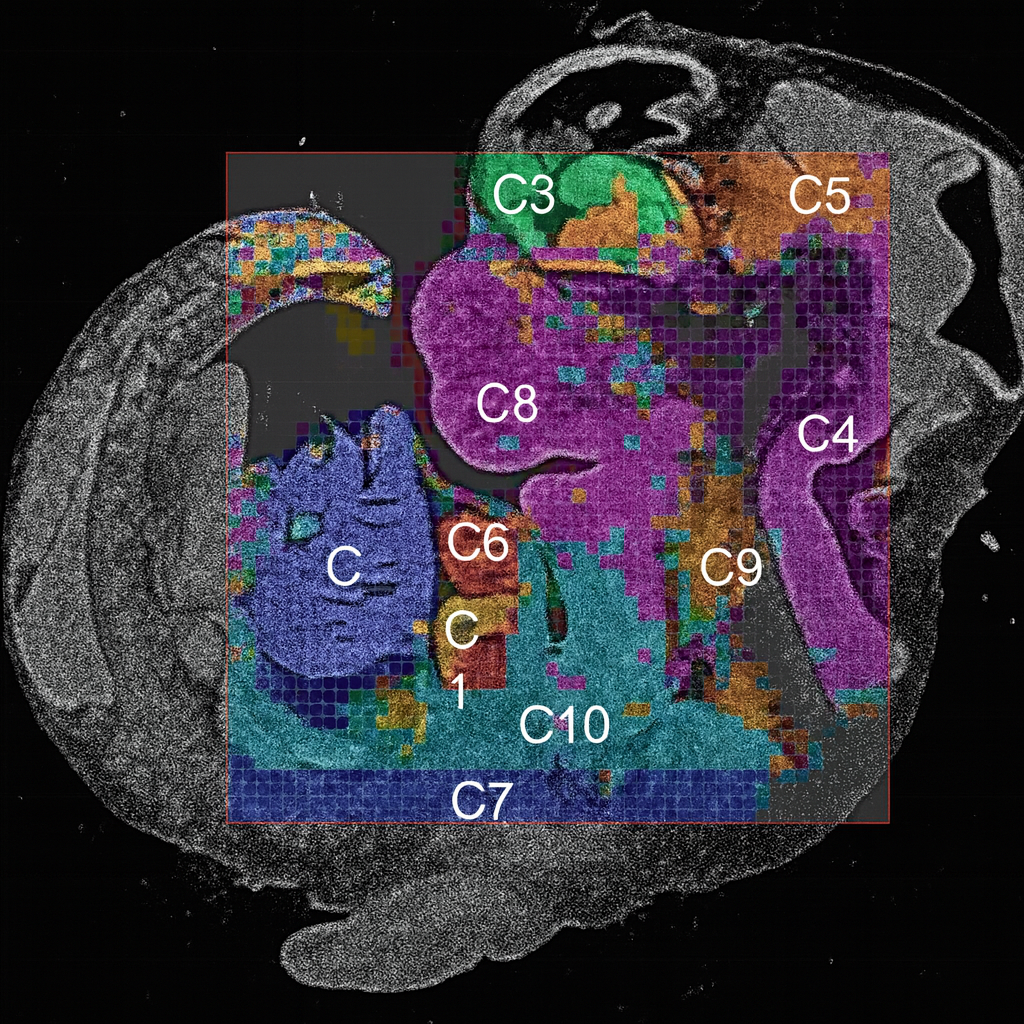
Spatial CUT&Tag
Map Histone Modifications and Chromatin-Bound Proteins
Spatial CUT&Tag for histone marks & chromatin-bound proteins
Antibody-guided Tn5 tagmentation maps chromatin modifications and protein–DNA occupancy directly on tissue sections, preserving morphology and neighborhood context.
-
✓
Targeted epigenomic readout
Profile activating, repressive, and architectural signals with spatial precision. -
✓
Gentle, streamlined workflow
On-slide binding and tagmentation reduce handling—well suited to delicate tissues and co-registration with H&E. -
✓
Quantitative spatial signals
Call peaks, compute enrichment, and relate programs to cell neighborhoods. -
✓
Complementary to Spatial ATAC
Layer CUT&Tag occupancy with accessibility to strengthen regulatory interpretation.
Spatial epigenomic profiling reveals the chromatin mechanisms driving tumor resistance and cellular heterogeneity. Using AtlasXomics’ spatial CUT&Tag and DBiT-seq platforms, we mapped histone modifications and chromatin accessibility across gastric and prostate tumors, uncovering group-specific enhancer activity, altered chromatin looping, and transcription factor networks linked to tumor-stroma interactions.

A deeper technical walkthrough of in-tissue, antibody-directed tagmentation for spatially resolved chromatin profiling, plus example results across marks and tissue contexts.
- How Spatial CUT&Tag works in situ (antibody-directed Tn5 tagmentation)
- Robust profiling across diverse chromatin targets
- Representative performance + reproducibility benchmarks

Contact Us
Have questions or interested in collaborating? We’d love to hear from you.
Get in Touch ↗