Our Platform

Our platform combines microfluidics and NGS to create multiomics maps
The method is composed of three main steps that apply to all of our spatial omics solutions and can be easily adapted to other next generation sequencing based assays.
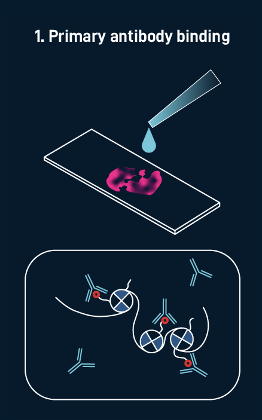
1. Labeling biomolecules in tissue

Primary antibody binding
Secondary antibody binding
pA-Tn5 transposition
The process begins with the addition of multi-omic probes (such as those for mRNA, genomic DNA, or proteins) that have a known sequence to a tissue section mounted on a standard pathology slide. In this CUT&TAg example, antibodies are added to direct the tagmentation process toward a specific histone modification. The resulting DNA fragments are then labeled with our custom adaptor, which is complementary to our spatial barcodes.
2. Spatial barcoding with microfluidic chips

Barcodes A ligation
Barcodes B ligation
Imaging
Sequentially and orthogonally applied microfluidic chips deliver x-barcodes and y-barcodes, that ligate to the known sequence, creating an in situ x-y coordinate system in tissue. Each intersection area between a microfluidic row and column generates a TIXEL,™ a square-shaped element of tissue.
3. Library preparation and sequencing

Reverse crosslinking and PCR
NGS Sequencing
Tissue is lysed and molecules labeled with our spatial barcodes are extracted for library preparation. Final library is sequenced and analyzed using standard single-cell bioinformatics pipelines.
The platform has been demonstrated across a wide range of tissue types
High reproducibility between serial sections
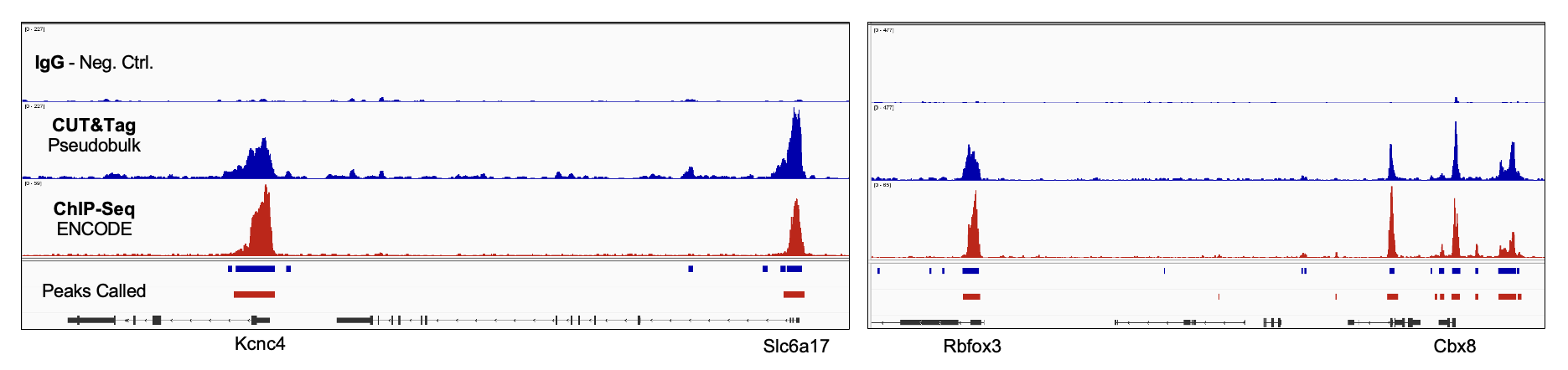
High sensitivity meets industry benchmarks
Strong correlation between data and peak calls for H3K4me3 pseudobulk spatial CUT&Tag and ChIP-Seq Reference data (ENCODE, Exp. ENCSR427ZJU). Kcnc4 and Rbfox3 loci pictured along with leg (Negative Control); Correlation between datasets was 0.78 (UCSC Browser utility, bigWigCorrelate.)